Revealing unexpected complex encoding but simple decoding mechanisms in motor cortex via separating behaviorally relevant neural signals
🔶 Understanding Neural Encoding and Decoding in Motor Cortex 🔶
🎯 Research Goal
Understanding how motor cortex (MC) encodes and decodes movement behaviors is a fundamental goal of neuroscience
(Kriegeskorte & Douglas, 2019; Saxena & Cunningham, 2019)
- Behavior = Measured task-specific behavioral variables (e.g., arm kinematics)
- Terms like “behaviorally relevant” or “irrelevant” refer only to these variables
⚠️ Key Challenge
❗ Neural signals are entangled:
- Relevant neural responses are mixed with:
- Irrelevant variables (Fusi et al., 2016; Rigotti et al., 2013)
- Ongoing neural noise (Azouz & Gray, 1999; Faisal et al., 2008)
🔍 This raises a critical question:
Could irrelevant signals conceal important facts about neural encoding and decoding?
🧩 Implications of Irrelevant Signals
🔬 Neural Encoding
- Irrelevant signals may:
- Mask small neural components
- Lead to underestimation of weakly tuned neurons
- Obscure informative low-variance PCs
Example Misconceptions:
- Weakly tuned neurons often ignored (Georgopoulos et al., 1986; Hochberg et al., 2012)
- Low-variance PCs discarded as noise (Churchland et al., 2012; Gallego et al., 2018, 2020)
❓ Do these components truly lack information, or are they just obscured?
🧠 Neural Decoding
- Irrelevant signals make information readout harder (Pitkow et al., 2015; Yang et al., 2021)
- Debate: Is motor cortex decoding:
- Linear (plausible and common)
- Nonlinear (performs better in practice, e.g., Glaser et al., 2020)
❓ Are irrelevant signals the reason nonlinear models outperform linear ones?
🧰 Proposed Solution: Signal Separation Framework
✅ Goal:
Separate behaviorally relevant and irrelevant signals at:
- Single-neuron
- Single-trial levels
🚧 Challenges:
- Ground truth of relevant signals is unknown
- Existing methods rely on assumptions:
- Focus on latent population (not single neurons)
- Assume specific dynamics (linear/nonlinear)
Limitations of Existing Approaches:
- Trial-averaged signals lose single-trial dynamics
- Assumption-based models miss out on nonconforming activity
🆕 Our Novel Framework
✨ Key Idea:
Define, extract, and validate behaviorally relevant signals based on:
- Encoding: Should resemble raw signals (preserve neural features)
- Decoding: Should retain maximal behavioral information
➡️ Establishes foundation for accurate mechanistic analysis
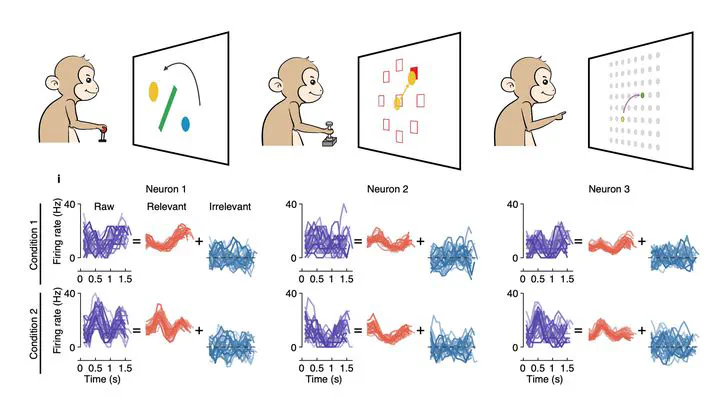
🧪 Experimental Setup
- Subjects: Three monkeys
- Task: Different reaching tasks
- Behavioral variable: Movement kinematics
🔍 Key Findings
1. Behaviorally Irrelevant Signals
- Explain a large portion of trial-to-trial variability
- Evenly distributed across dimensions of relevant signals
2. Neural Encoding Insights
- Irrelevant signals obscure behaviorally meaningful activity
- Responses with nonlinear encoding are especially affected
🚫 Previously Ignored Signals:
- Weakly tuned neurons
- Low-variance PCs
✅ Actually encode rich behavioral information in nonlinear ways
- Reveal neuronal redundancy
- Suggest a higher-dimensional neural representation of movement
🧠 Synergistic Effect:
- Combining small & large variance PCs improves decoding
- Small variance PCs enhance fine-grained speed control → Crucial for precise motor control
3. Neural Decoding Insights
- Irrelevant signals complicate decoding
- Linear decoders can match nonlinear decoders once irrelevant signals are removed → Supports the possibility of linear readout in motor cortex
✅ Encoding = Complex
✅ Decoding = Simple
🚀 Broader Impact
🔧 Implications for:
- Brain-machine interfaces (BMIs): More accurate & robust decoding
- Systems neuroscience: General framework to separate signals and uncover hidden neural mechanisms across cortical areas
📚 Key References
- Kriegeskorte & Douglas, 2019
- Saxena & Cunningham, 2019
- Fusi et al., 2016; Rigotti et al., 2013
- Azouz & Gray, 1999; Faisal et al., 2008
- Georgopoulos et al., 1986; Hochberg et al., 2012; Wodlinger et al., 2015
- Churchland et al., 2012; Gallego et al., 2018, 2020
- Pitkow et al., 2015; Yang et al., 2021
- Glaser et al., 2020; Willsey et al., 2022
- Sani et al., 2021; Hurwitz, 2021; Zhou, 2020
- Kobak et al., 2016; Rouse & Schieber, 2018